A temperature-tolerant CRISPR base editor mediates highly efficient and precise gene inactivation in vivo
Abstract
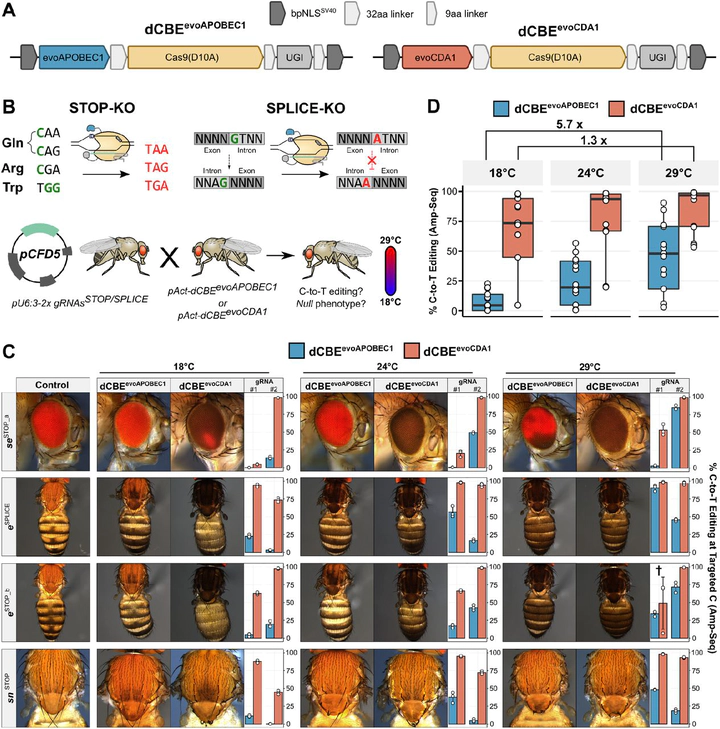
CRISPR nucleases generate a broad spectrum of mutations that includes undesired editing outcomes which attenuate phenotypes and complicate experimental analysis and interpretation. Here, we develop an optimised cytosine base editing system for gene inactivation in Drosophila through predictable C-to-T editing and identify temperature as a crucial parameter for base editing efficiency. We find that activity of an evolved version of the most widely used APOBEC1 deaminase is attenuated within the temperature range commonly used for culturing Drosophila (18-29°C) and many other ectothermic species. In contrast, an evolved CDA1 domain functions with remarkable efficiency within the same temperature range. Furthermore, we show that formation of undesired indel mutations and C-to-G/A edits is exceptionally rare in Drosophila compared to other species. The predictable editing outcome, very high efficiency and minimal byproduct formation of this system allows for near homogeneous biallelic gene inactivation in vivo in a ubiquitous or conditional manner. This work significantly improves our ability to create precise loss-of-function alleles in Drosophila and provides key design parameters for developing highly efficient base editing systems in other ectothermic species.