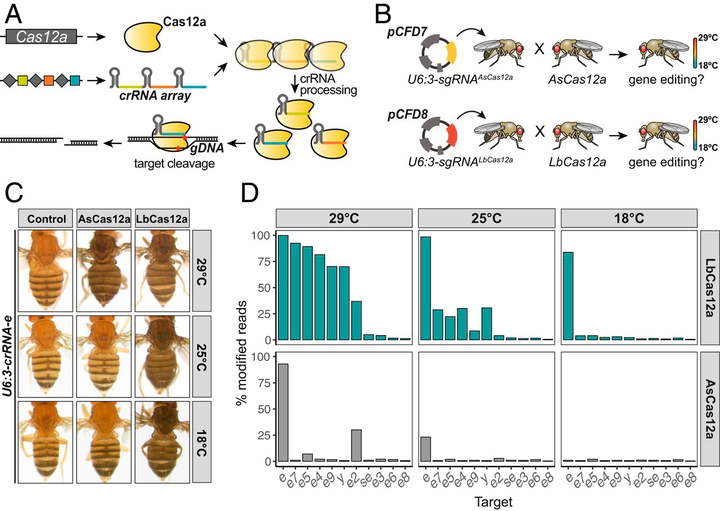
Abstract
CRISPR-Cas genome engineering has revolutionized biomedical research by enabling targeted genome modification with unprecedented ease. In the popular model organism Drosophila melanogaster, gene editing has so far relied exclusively on the prototypical CRISPR nuclease Cas9. Additional CRISPR systems could expand the genomic target space, offer additional modes of regulation, and enable the independent manipulation of genes in different cells of the same animal. Here we describe a platform for efficient Cas12a gene editing in Drosophila. We show that Cas12a from Lachnospiraceae bacterium, but not Acidaminococcus spec., can mediate robust gene editing in vivo. In combination with most CRISPR RNAs (crRNAs), LbCas12a activity is high at 29 °C, but low at 18 °C, enabling modulation of gene editing by temperature. LbCas12a can directly utilize compact crRNA arrays that are substantially easier to construct than Cas9 single-guide RNA arrays, facilitating multiplex genome engineering. Furthermore, we show that conditional expression of LbCas12a is sufficient to mediate tightly controlled gene editing in a variety of tissues, allowing detailed analysis of gene function in a multicellular organism. We also test a variant of LbCas12a with a D156R point mutation and show that it has substantially higher activity and outperforms a state-of-the-art Cas9 system in identifying essential genes. Cas12a gene editing expands the genome-engineering toolbox in Drosophila and will be a powerful method for the functional annotation of the genome. This work also presents a fully genetically encoded Cas12a system in an animal, laying out principles for the development of similar systems in other genetically tractable organisms for multiplexed conditional genome engineering.